Research
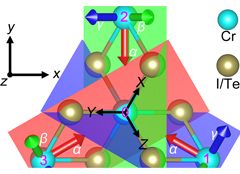
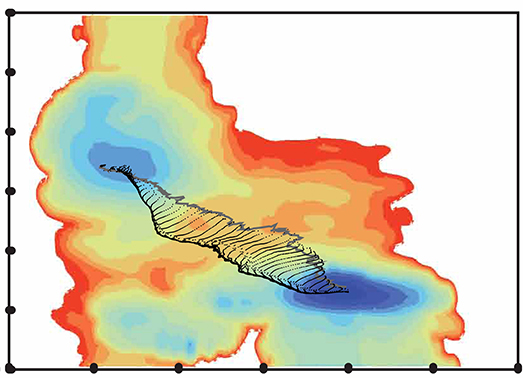
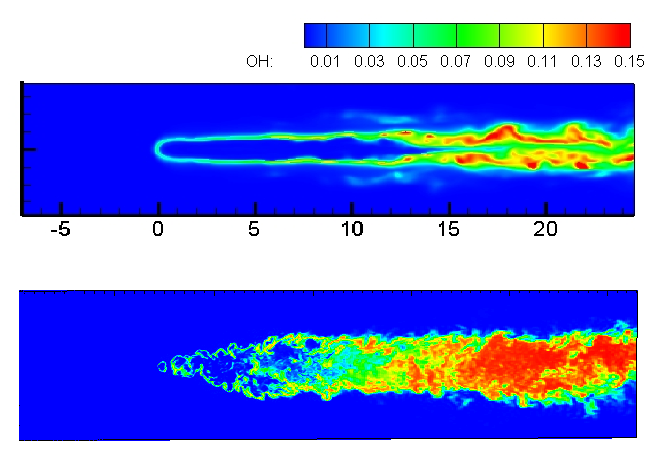
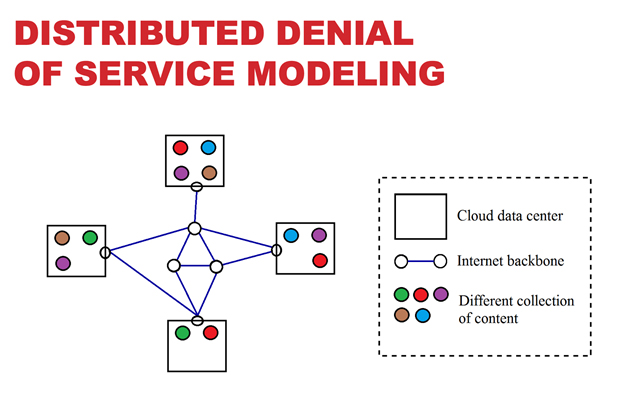
University of Arkansas researchers use the AHPCC core research facility to enable computationally intensive research across many disciplines. Below are examples of many research areas which AHPCC staff and equipment provide support for. While not inclusive, the examples displayed represent institutional areas which frequently utilize large portions of AHPCC resources to advance the mission of research to provide significant discovery, insight, and understanding.