Chemistry
The Biomolecular Simulations Group (the Moradi Lab) at the University of Arkansas use high performance computing to simulate the conformational changes of proteins at an atomic level. Moradi's research helps shed light on the structure function relationships in proteins and can help improve our understanding of disease at a molecular level. Visit the Biomolecular Simulations Group for more information on their current projects and for a complete publications list.
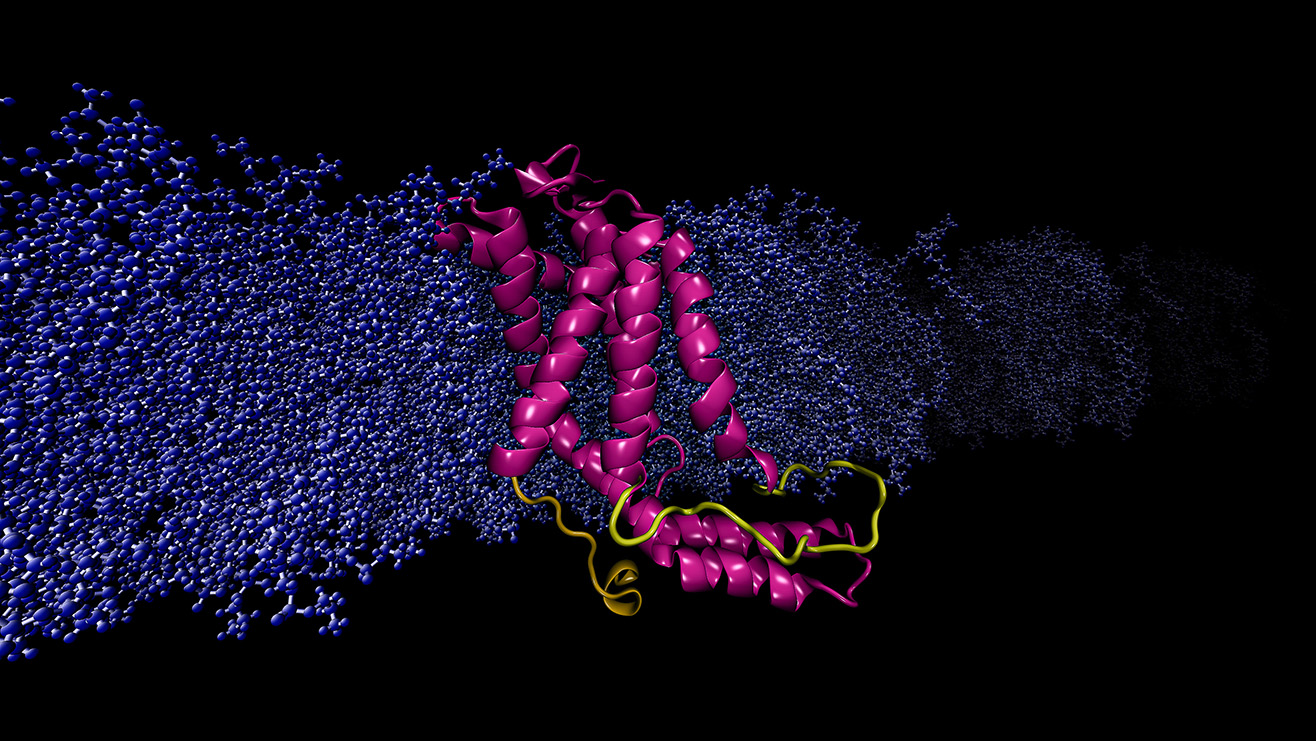 Figure 1: The modeled structure of bacterial membrane insertase protein YidC2 based
on microsecond level simulations of membrane embedded protein using AHPCC resources.
Figure 1: The modeled structure of bacterial membrane insertase protein YidC2 based
on microsecond level simulations of membrane embedded protein using AHPCC resources.
The Predictive Compuational Chemistry Laboratory (Feng Wang group) researches basic fundamental properties of potential drug candidates, such as solubility
and binding affinity. With the new Pinnacle cluster at the AHPCC, large scale parallel
electronic structure computation are performed to provide information needed to fit
accurate models for drug molecules. Other projects include the development of models
for cellulose, the main component of biomass.
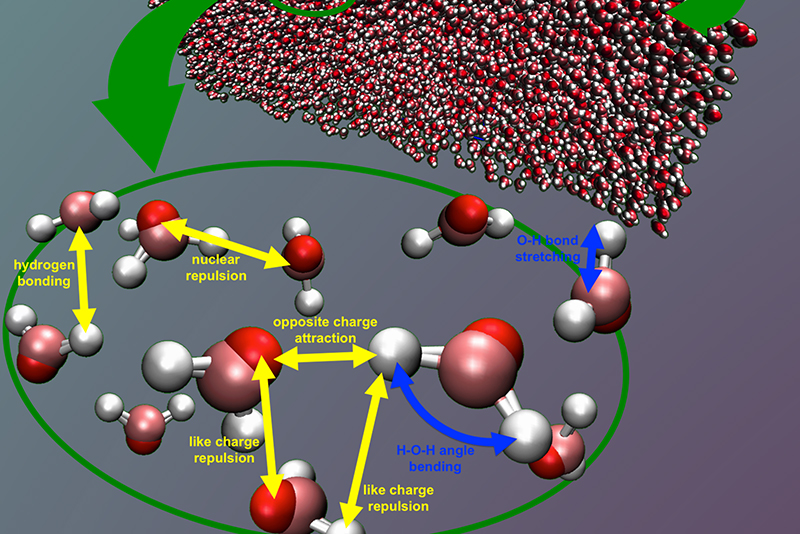 Figure 2: Using a very accurate water model, Trestles was used to perform high quality
computer measurements of liquid water’s surface tension in deep supercooling. This
study predicted a large increase in the surface tension of water around -38 °C. Our
study explains a small divergence observed experimentally in less supercooled water.
This large rise in the surface tension supports the existence of a second liquid phase
of water, which has been debated for many years.
Figure 2: Using a very accurate water model, Trestles was used to perform high quality
computer measurements of liquid water’s surface tension in deep supercooling. This
study predicted a large increase in the surface tension of water around -38 °C. Our
study explains a small divergence observed experimentally in less supercooled water.
This large rise in the surface tension supports the existence of a second liquid phase
of water, which has been debated for many years.